Patient-Derived Xenografts, the Cancer Stem Cell Paradigm, and Cancer Pathobiology in the 21st Centu
Patient-Derived Xenografts, the Cancer Stem Cell Paradigm, and Cancer Pathobiology in the 21st Century
More than 68 drugs with varying efficacy have been developed and approved for oncology over the last several decades, yet the unfortunate paucity of success stories serves as a reminder that the current arsenal of therapies fail far too many patients. It is tempting to speculate that failure of current therapies stems from the limitations of the tissue culture model systems in which they were discovered, validated and/or evaluated for potency during preclinical development. In the late 1970's, colony-forming soft agar assays emerged as a means by which scientists attempted to study the nature and potential of human tumor stem cells. These in vitro colony-forming assays have facilitated insight into the differentiation potential and relationship of hematopoietic stem and progenitor cell populations in normal hematopoiesis and hematopoietic malignancies; however, little practical insight has been garnered in solid tissues and tumors.
Traditional cell lines originally derived from patient tumors and adapted to proliferate in in vitro culture conditions have been widely established and studied for more than a half century. These lines have served as a foundation for cancer research due to their ability to be easily propagated and studied under defined conditions. Unfortunately, continuous passage and culture of cells in vitro tends to select for cells adapted to thrive in plastic dishes and eliminates variables introduced by tumor resident cell populations such as supporting non-tumor stroma, hematopoietic cells and other tumor microenvironmental factors such as extracellular matrix proteins. In conjunction with the harsh enzymatic manipulations and centrifugation conditions employed during the passage of traditional cell lines, in vitro culture conditions have selected for cellular subsets that flourish in their newfound laboratory setting, and have generally selected for populations that are phenotypically uniform—a gross departure from the natural tumor state.
Traditional cell lines are maintained in culture conditions that depart markedly from the natural setting of human tumors. Specifically, cell lines are cultured in nutrient-rich media in high oxygen tension as plastic-adherent monolayers or in suspension with a lack of any attachment substrate. Not only are these cell lines typically clonal, but they are generally inefficient at initiating tumors when transplanted into compatible, immunocompromised hosts such as nude, SCID, NOD/SCID, or NOD/SCID/γ-null (ie, NSG) mice. When tumors do arise, these masses are generally homogeneous in nature and do not reflect tumor biology as it exists in patients (eg, Figures 1a and b). The expansion of cells in aphysiologic oxygen concentrations (ie, 21% O2: ambient oxygen levels) has contributed to the perpetuation and study of cells that have accrued dozens, if not hundreds, of mutations and chromosomal abnormalities over the course of their many passages in vitro. Profound karyotypic dysmorphims, including hypotriploid genotypes, are surprisingly common within the cell lines that have come to define the standard models studied by many cancer biologists, such as the NCI60 cell lines (also see http://www.ncbi.nlm.nih.gov/sky/skyweb.cgi). Recent studies have demonstrated that brief periods of in vitro culture irreversibly change gene expression, indicating that even low-passage cell lines may be compromised. Collectively, these observations call into question the physiological relevance of traditional cell lines as sufficient, let alone relevant, models for studying tumor biology as it exists in cancer patients.
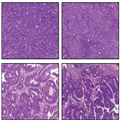
(Enlarge Image)
Figure 1.
Patient-derived xenograft (PDX) models of colorectal cancer recapitulate primary tumor heterogeneity. Hematoxylin and eosin (H&E) stained FFPE slides of xenografts generated by traditional HT-29 (a) or SW480 (b) colorectal cancer cell lines, vs a primary colorectal tumor, SCRX-PDX-CR101-p0 (c), and the same patient's PDX tumor following passaging through NOD/SCID mice, SCRX-PDX-CR101-p1 (d). Note the relative uniformity of the HT-29 and SW480 tumors relative to the primary or PDX tumor following minimal passaging in immunocompromised mice.
Despite the increasingly apparent limitations of traditional cell lines, they have made significant contributions to our understanding of tumor biology. The uniformity and control over experimental conditions afforded by in vitro cell culture has assisted the development and execution of highly reproducible studies under defined conditions that enable insight into drug sensitivity, basic cell biology, and the elucidation of signaling pathways. For example, in vitro cell culture has facilitated the development of high-throughput approaches leading to the discovery of second and third generation anti-neoplastic agents (see DeVita and Chu for review). The flexibility and accessibility of traditional cell lines perpetuated in vitro has also enabled key mechanistic studies to shed light on the contribution of specific genes and mutations to cell survival, proliferation, and migration, contributing to the development of, for example, kinase-specific inhibitors such as erlotinib, vemurafenib, and crizotinib targeting EGFR, BRAF, and ALK/c-MET, respectively (reviewed by Sawyers). Although initially promising, these tailored kinase inhibitors have largely failed to be curative and typically extend life only 3–6 months. Although chemotherapeutic compounds have also proven highly effective in vitro, in traditional in vivo tumor models and often in the clinic, the untargeted nature of these drugs results in significant toxicity, and consequently, a narrow therapeutic window in cancer patients. Moreover, residual tumors generally recur as more aggressive, refractory, and lethal; likely due to the inability of chemotherapy to eliminate CSC, and resulting in additional mutations accumulated in these cells during exposure to genotoxic drug regimens.
Genetically engineered mouse models (GEMMs) are also popular models through which tumor biology is studied. Unfortunately, these tumor models also have their intrinsic shortcomings. It is becoming increasingly clear that more than one driver mutation is needed to initiate tumorigenesis—both in mice and in men. Efficient tumorigenesis in GEMMs must often be driven by the introduction of at least two defined oncogenes and/or mutated tumor suppressors, depending on the mouse model and/or models needed to be crossed to drive tumorigenesis. Generating GEMMs with more than one driver mutation is extremely time-consuming and difficult. Another weakness of these models is that transgene expression (eg, mutated KRAS) is commonly driven to superphysiological levels, such that protein expression often exceeds that ever encountered in patients. Moreover, when tumors do arise in these models, they are sporadic in their growth such that it is difficult to design studies powered by a significant number of animals, while also being difficult to monitor without labor-intensive in vivo imaging technologies. As previously discussed, the Cancer Genome Atlas project has demonstrated that the spectrum of driver mutations differs significantly among patients, thus the relevance of particular GEMMs to patients in the clinic is increasingly questionable. Notwithstanding the above criticisms, GEMMs may have advantages over xenograft models in their cellular composition and/or sensitivity to therapeutic agents. Specifically, GEMM tumors contain stromal and hematopoietic components not possible in a human tumor xenograft setting, and thus these tumors may respond more appropriately to small molecules, immunomodulatory, and/or biological agents that cross-react with mouse antigens. Nevertheless, for the various reasons outlined above, GEMMs will likely provide limited insight into oncogenesis and patient tumor heterogeneity.
Although traditional cell lines have helped gain an understanding of the basic biology underlying specific genes, proteins, and/or signaling pathways, cell lines cultured in vitro have not significantly contributed to the discovery of targets for directed therapies that have meaningfully impacted patient survival. Trastuzumab and T-DM1 (anti-Her2 biologics) and Imatinib/Gleevec (a BCR/ABL kinase inhibitor) are prime examples of cancer drugs that have meaningfully impacted patient survival; however, neither was discovered in traditional cell lines or GEMMs. Her2/Neu (ErbB2) was originally identified as a potentially interesting cancer target based on the observation of elevated expression in tumor specimens obtained and preserved by pathologists. Moreover, the p210-BCR/ABL fusion protein was identified and associated with chronic myelogenous leukemia (CML) upon close examination of chromosomal architecture using blood smears from CML patients. Traditional cell lines and GEMMs effectively complemented these discoveries, but were most effectively utilized as platforms to precisely dissect the molecular and cellular biology of ErbB2 and p210-BCR/ABL, respectively. To understand tumorigenesis and patient heterogeneity observed in the clinic, cancer research, drug discovery, and development must increasingly look beyond in vitro tissue culture models and GEMMs and towards experimental systems that better replicate human tumor biology and enable the study of many patient tumors as they are likely to exist in cancer patients.
Traditional Models Have Not Predicted Clinical Success
More than 68 drugs with varying efficacy have been developed and approved for oncology over the last several decades, yet the unfortunate paucity of success stories serves as a reminder that the current arsenal of therapies fail far too many patients. It is tempting to speculate that failure of current therapies stems from the limitations of the tissue culture model systems in which they were discovered, validated and/or evaluated for potency during preclinical development. In the late 1970's, colony-forming soft agar assays emerged as a means by which scientists attempted to study the nature and potential of human tumor stem cells. These in vitro colony-forming assays have facilitated insight into the differentiation potential and relationship of hematopoietic stem and progenitor cell populations in normal hematopoiesis and hematopoietic malignancies; however, little practical insight has been garnered in solid tissues and tumors.
Traditional cell lines originally derived from patient tumors and adapted to proliferate in in vitro culture conditions have been widely established and studied for more than a half century. These lines have served as a foundation for cancer research due to their ability to be easily propagated and studied under defined conditions. Unfortunately, continuous passage and culture of cells in vitro tends to select for cells adapted to thrive in plastic dishes and eliminates variables introduced by tumor resident cell populations such as supporting non-tumor stroma, hematopoietic cells and other tumor microenvironmental factors such as extracellular matrix proteins. In conjunction with the harsh enzymatic manipulations and centrifugation conditions employed during the passage of traditional cell lines, in vitro culture conditions have selected for cellular subsets that flourish in their newfound laboratory setting, and have generally selected for populations that are phenotypically uniform—a gross departure from the natural tumor state.
Traditional cell lines are maintained in culture conditions that depart markedly from the natural setting of human tumors. Specifically, cell lines are cultured in nutrient-rich media in high oxygen tension as plastic-adherent monolayers or in suspension with a lack of any attachment substrate. Not only are these cell lines typically clonal, but they are generally inefficient at initiating tumors when transplanted into compatible, immunocompromised hosts such as nude, SCID, NOD/SCID, or NOD/SCID/γ-null (ie, NSG) mice. When tumors do arise, these masses are generally homogeneous in nature and do not reflect tumor biology as it exists in patients (eg, Figures 1a and b). The expansion of cells in aphysiologic oxygen concentrations (ie, 21% O2: ambient oxygen levels) has contributed to the perpetuation and study of cells that have accrued dozens, if not hundreds, of mutations and chromosomal abnormalities over the course of their many passages in vitro. Profound karyotypic dysmorphims, including hypotriploid genotypes, are surprisingly common within the cell lines that have come to define the standard models studied by many cancer biologists, such as the NCI60 cell lines (also see http://www.ncbi.nlm.nih.gov/sky/skyweb.cgi). Recent studies have demonstrated that brief periods of in vitro culture irreversibly change gene expression, indicating that even low-passage cell lines may be compromised. Collectively, these observations call into question the physiological relevance of traditional cell lines as sufficient, let alone relevant, models for studying tumor biology as it exists in cancer patients.

(Enlarge Image)
Figure 1.
Patient-derived xenograft (PDX) models of colorectal cancer recapitulate primary tumor heterogeneity. Hematoxylin and eosin (H&E) stained FFPE slides of xenografts generated by traditional HT-29 (a) or SW480 (b) colorectal cancer cell lines, vs a primary colorectal tumor, SCRX-PDX-CR101-p0 (c), and the same patient's PDX tumor following passaging through NOD/SCID mice, SCRX-PDX-CR101-p1 (d). Note the relative uniformity of the HT-29 and SW480 tumors relative to the primary or PDX tumor following minimal passaging in immunocompromised mice.
Despite the increasingly apparent limitations of traditional cell lines, they have made significant contributions to our understanding of tumor biology. The uniformity and control over experimental conditions afforded by in vitro cell culture has assisted the development and execution of highly reproducible studies under defined conditions that enable insight into drug sensitivity, basic cell biology, and the elucidation of signaling pathways. For example, in vitro cell culture has facilitated the development of high-throughput approaches leading to the discovery of second and third generation anti-neoplastic agents (see DeVita and Chu for review). The flexibility and accessibility of traditional cell lines perpetuated in vitro has also enabled key mechanistic studies to shed light on the contribution of specific genes and mutations to cell survival, proliferation, and migration, contributing to the development of, for example, kinase-specific inhibitors such as erlotinib, vemurafenib, and crizotinib targeting EGFR, BRAF, and ALK/c-MET, respectively (reviewed by Sawyers). Although initially promising, these tailored kinase inhibitors have largely failed to be curative and typically extend life only 3–6 months. Although chemotherapeutic compounds have also proven highly effective in vitro, in traditional in vivo tumor models and often in the clinic, the untargeted nature of these drugs results in significant toxicity, and consequently, a narrow therapeutic window in cancer patients. Moreover, residual tumors generally recur as more aggressive, refractory, and lethal; likely due to the inability of chemotherapy to eliminate CSC, and resulting in additional mutations accumulated in these cells during exposure to genotoxic drug regimens.
Genetically engineered mouse models (GEMMs) are also popular models through which tumor biology is studied. Unfortunately, these tumor models also have their intrinsic shortcomings. It is becoming increasingly clear that more than one driver mutation is needed to initiate tumorigenesis—both in mice and in men. Efficient tumorigenesis in GEMMs must often be driven by the introduction of at least two defined oncogenes and/or mutated tumor suppressors, depending on the mouse model and/or models needed to be crossed to drive tumorigenesis. Generating GEMMs with more than one driver mutation is extremely time-consuming and difficult. Another weakness of these models is that transgene expression (eg, mutated KRAS) is commonly driven to superphysiological levels, such that protein expression often exceeds that ever encountered in patients. Moreover, when tumors do arise in these models, they are sporadic in their growth such that it is difficult to design studies powered by a significant number of animals, while also being difficult to monitor without labor-intensive in vivo imaging technologies. As previously discussed, the Cancer Genome Atlas project has demonstrated that the spectrum of driver mutations differs significantly among patients, thus the relevance of particular GEMMs to patients in the clinic is increasingly questionable. Notwithstanding the above criticisms, GEMMs may have advantages over xenograft models in their cellular composition and/or sensitivity to therapeutic agents. Specifically, GEMM tumors contain stromal and hematopoietic components not possible in a human tumor xenograft setting, and thus these tumors may respond more appropriately to small molecules, immunomodulatory, and/or biological agents that cross-react with mouse antigens. Nevertheless, for the various reasons outlined above, GEMMs will likely provide limited insight into oncogenesis and patient tumor heterogeneity.
Although traditional cell lines have helped gain an understanding of the basic biology underlying specific genes, proteins, and/or signaling pathways, cell lines cultured in vitro have not significantly contributed to the discovery of targets for directed therapies that have meaningfully impacted patient survival. Trastuzumab and T-DM1 (anti-Her2 biologics) and Imatinib/Gleevec (a BCR/ABL kinase inhibitor) are prime examples of cancer drugs that have meaningfully impacted patient survival; however, neither was discovered in traditional cell lines or GEMMs. Her2/Neu (ErbB2) was originally identified as a potentially interesting cancer target based on the observation of elevated expression in tumor specimens obtained and preserved by pathologists. Moreover, the p210-BCR/ABL fusion protein was identified and associated with chronic myelogenous leukemia (CML) upon close examination of chromosomal architecture using blood smears from CML patients. Traditional cell lines and GEMMs effectively complemented these discoveries, but were most effectively utilized as platforms to precisely dissect the molecular and cellular biology of ErbB2 and p210-BCR/ABL, respectively. To understand tumorigenesis and patient heterogeneity observed in the clinic, cancer research, drug discovery, and development must increasingly look beyond in vitro tissue culture models and GEMMs and towards experimental systems that better replicate human tumor biology and enable the study of many patient tumors as they are likely to exist in cancer patients.
